AFM Systems
AFM Accessories
Learning
Contact Us
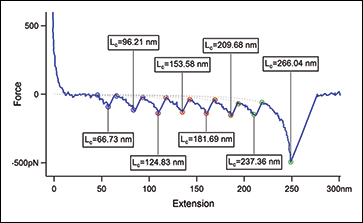
Atomic force microscopy is a powerful tool capable of measuring the mechanics of materials, ranging from soft biomaterial (molecules, cells, tissues, etc.) to polymers and harder inorganics. Depending on the spring constant of the cantilevers, pN-scale forces can be measured as either an individual molecule is unfolded (intramolecular) or two molecules are pulled apart (intermolecular).
Ask an AFM expert for more information"Angle-dependent atomic force microscopy single-chain pulling of adsorbed macromolecules from planar surfaces unveils the signature of an adsorption–desorption transition," L. Grebíková, S. G. Whittington, and J. G. Vancso, J. Am. Chem. Soc. 140, 6408 (2018). https://doi.org/10.1021/jacs.8b02851
"Dynamic and depth dependent nanomechanical properties of dorsal ruffles in live cells and biopolymeric hydrogels," V. Vyas, M. Solomon, G. G. D'Souza, and B. D. Huey, J. Nanosci. Nanotechnol. 18, 1557 (2018). https://doi.org/10.1166/jnn.2018.14219
"Measuring nanoscale viscoelastic parameters of cells directly from AFM force-displacement curves," Y. M. Efremov, W.-H. Wang, S. D. Hardy, R. L. Geahlen, and A. Raman, Sci. Rep. 7, 1541 (2017). https://doi.org/10.1038/s41598-017-01784-3
"Mechanical properties of membranes composed of gel-phase or fluid-phase phospholipids probed on liposomes by atomic force spectroscopy," O. Et-Thakafy, N. Delorme, C. Gaillard, C. Mériadec, F. Artzner, C. Lopez, and F. Guyomarc'h, Langmuir 33, 5117 (2017). https://doi.org/10.1021/acs.langmuir.7b00363
"Atomic force microscopy-guided fractionation reveals the influence of cranberry phytochemicals on adhesion of Escherichia coli," P. Gupta, B. Song, C. Neto, and T. A. Camesano, Food Funct. 7, 2655 (2016). https://doi.org/10.1039/c6fo00109b
"Correlating confocal microscopy and atomic force indentation reveals metastatic cancer cells stiffen during invasion into collagen I matrices," J. R. Staunton, B. L. Doss, S. Lindsay, and R. Ros, Sci. Rep. 6, 19686 (2016). https://doi.org/10.1038/srep19686
"Adsorption and adhesion of common serum proteins to nanotextured gallium nitride," L. E. Bain, M. P. Hoffmann, I. Bryan, R. Collazo, and A. Ivanisevic, Nanoscale 7, 2360 (2015). hhttps://doi.org/10.1039/c4nr06353h
"Toughness and damage susceptibility in human cortical bone is proportional to mechanical inhomogeneity at the osteonal-level," O. L. Katsamenis, T. Jenkins, and P. J. Thurner, Bone 76, 158 (2015). http://dx.doi.org/10.1016/j.bone.2015.03.020
"Force measurements on natural membrane nanovesicles reveal a composition-independent, high Young's modulus," A. Calò, D. Reguera, G. Oncins, M.-A. Persuy, G. Sanz, S. Lobasso, A. Corcelli, E. Pajot-Augy, and G. Gomila, Nanoscale 6, 2275 (2014). https://doi.org/10.1039/c3nr05107b
"Probing the hydrophobic interaction between air bubbles and partially hydrophobic surfaces using atomic force microscopy," C. Shi, D. Y. Chan, Q. Liu, and H. Zeng, J. Phys. Chem. C 118, 25000 (2014). https://doi.org/10.1021/jp507164c
"Measuring the interaction between ions, biopolymers and interfaces — one polymer at a time," S. Kienle, T. Pirzer, S. Krysiak, M. Geisler, and T. Hugel, Faraday Discuss. 160, 329 (2013). https://doi.org/10.1039/c2fd20069d
"Measurement of the hydrophobic force in a soft matter system," R. F. Tabor, C. Wu, F. Grieser, R. R. Dagastine, and D. Y. C. Chan, J. Phys. Chem. Lett. 4, 3872 (2013). https://doi.org/10.1021/jz402068k
"Poly(acrylamide) films at the solvent-induced glass transition: adhesion, tribology, and the influence of crosslinking," A. Li, S. N. Ramakrishna, E. S. Kooij, R. M. Espinosa-Marzal, and N. D. Spencer, Soft Matter 8, 9092 (2012). https://doi.org/10.1039/c2sm26222c
"Thickness-corrected model for nanoindentation of thin films with conical indenters," J. A. C. Santos, L. M. Rebêlo, A. C. Araujo, E. B. Barros, and J. S. de Sousa, Soft Matter 8, 4441 (2012). https://doi.org/10.1039/c2sm07062f
"Evolutionary screening of collagen-like peptides that nucleate hydroxyapatite crystals," W.-J. Chung, K.-Y. Kwon, J. Song, and S.-W. Lee, Langmuir 27, 7620 (2011). https://doi.org/10.1021/la104757g
"Single-molecule determination of the face-specific adsorption of Amelogenin's C-terminus on hydroxyapatite," R. W. Friddle, K. Battle, V. Trubetskoy, J. Tao, E. A. Salter, J. Moradian-Oldak, J. J. De Yoreo, and A. Wierzbicki, Angew. Chem. Int. Ed. 50, 7541 (2011). https://doi.org/10.1002/anie.201100181
"Molecular shape and binding force of Mycoplasma mobile's leg protein Gli349 revealed by an AFM study," C. Lesoil, T. Nonaka, H. Sekiguchi, T. Osada, M. Miyata, R. Afrin, and A. Ikai, Biochem. Biophys. Res. Commun. 391, 1312 (2010). https://doi.org/10.1016/j.bbrc.2009.12.023
"Elastic modulus of single cellulose microfibrils from tunicate measured by atomic force microscopy," S. Iwamoto, W. Kai, A. Isogai, and T. Iwata, Biomacromolecules 10, 2571 (2009). https://doi.org/10.1021/bm900520n
"Single-molecule measurement of the strength of a siloxane bond," P. Schwaderer, E. Funk, F. Achenbach, J. Weis, C. Bräuchle, and J. Michaelis, Langmuir 24, 1343 (2008). https://doi.org/10.1021/la702352x
"A reversible wet/dry adhesive inspired by mussels and geckos," H. Lee, B. P. Lee, and P B. Messersmith, Nature 448, 338 (2007). https://doi.org/10.1038/nature05968
"Direct force measurements between carboxylate-modified latex microspheres and glass using atomic force microscopy," S. Assemi, J. Nalaskowski, and W. P. Johnson, Colloids Surf. A 286, 70 (2006). https://doi.org/10.1016/j.colsurfa.2006.03.024
"Sacrificial bonds in polymer brushes from rat tail tendon functioning as nanoscale velcro," T. Gutsmann, T. Hassenkam, J. A. Cutroni, and P. K. Hansma, Biophys. J. 89, 536 (2005). https://doi.org/10.1529/biophysj.104.056747
"Surface probe measurements of the elasticity of sectioned tissue, thin gels and polyelectrolyte multilayer films: Correlations between substrate stiffness and cell adhesion," A. J. Engler, L. Richert, J. Y. Wong, C. Picart, and D .E. Discher, Surf. Sci. 570, 142 (2004). https://doi.org/10.1016/j.susc.2004.06.179
"The biomechanics toolbox: experimental approaches for living cells and biomolecules," K. V. Vliet, G. Bao, and S. Suresh, Acta Mater. 51, 5881 (2003). https://doi.org/10.1016/j.actamat.2003.09.001
"Determination of elastic moduli of thin layers of soft material using the atomic force microscope," E. K. Dimitriadis, F. Horkay, J. Maresca, B. Kachar, and R. S. Chadwick, Biophys. J. 82, 2798 (2002). https://doi.org/10.1016/s0006-3495(02)75620-8