AFM Systems
AFM Accessories
Learning
Contact Us
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
High-resolution AFM imaging in ionic aqueous solution gave detailed insight into the structure and formation of DNA G-wires. The observed nanoscale periodic features allowed researchers to classify the G-wires into different species.
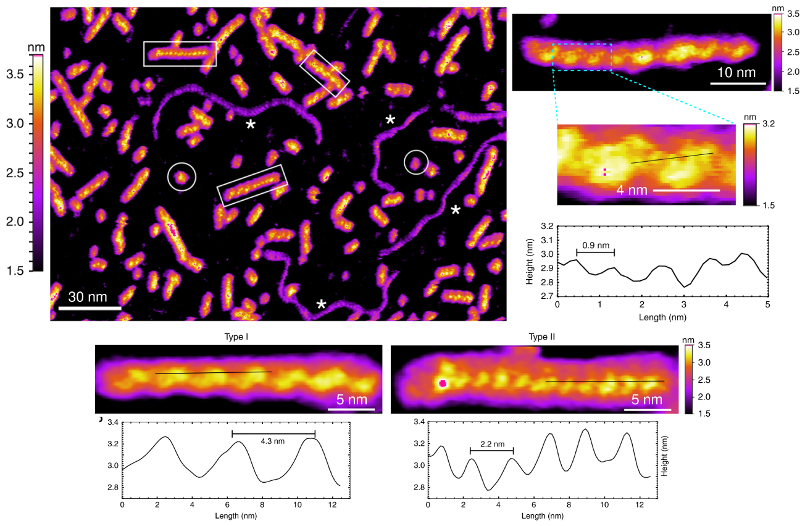
G-wire DNA is a family of nucleic acid structures formed by self-assembly of short DNA fragments. These wire-like structures have attracted interest not only for their biological implications but also for potential use in nanoelectronic and nanosensor devices. However, the details of how G-wires form and the biomolecular structures that result are still not clearly understood.
To this end, researchers from Singapore’s Nanyang Technological University investigated self-assembly of short pieces of the Tetrahymena telomeric DNA sequence d[G4T2G4]. They acquired high-resolution AFM images in physiologically relevant conditions and observed a variety of wire-like structures with periodic features as small as 0.9 nm.
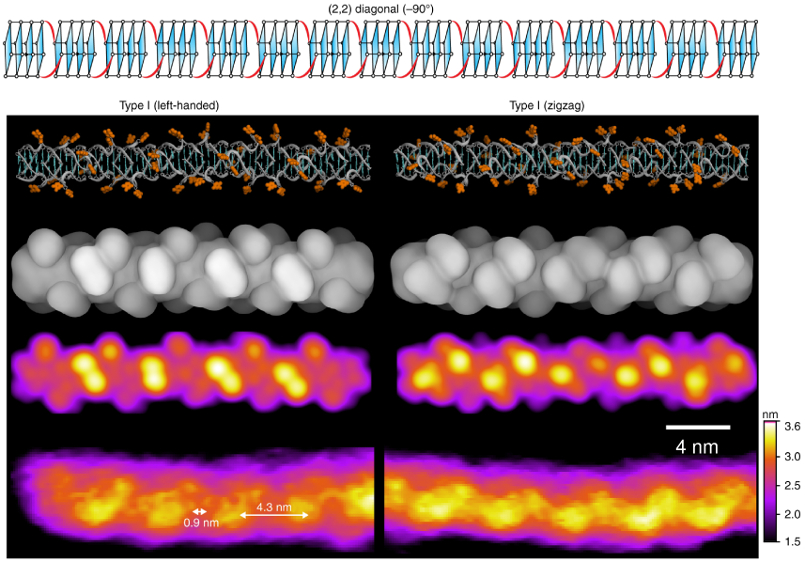
From this data, the researchers classified G-wires into different species according to height, periodicity, and handedness. Molecular modeling and simulations also allowed them to identify a specific molecular structure (rotamer) that could explain the experimental observations.
By enriching understanding of G-wire formation, the results may prove valuable for a range of bio- and nanotechnology applications related to DNA self-assembly.
Cypher ES with Heating and Cooling Sample Stage
G-wires immersed in potassium phosphate buffer solution were imaged on the Cypher ES AFM in tapping mode. Experiments were performed with the solution cooled to a constant 10 °C using the Heating and Cooling Sample Stage. The nanoscale structures were clearly visualized due to the unmatched spatial resolution routinely achieved by Cypher AFMs. In addition, blueDrive technology helped make tapping in liquid simpler and more stable. The G-wires’ nanomechanical properties, including stiffness and loss tangent, were characterized with AM-FM Viscoelastic Mapping Mode (see supplementary information available at the article link below).
Citation: K. Bose, C. J. Lech, B. Heddi, and A. T. Phan, High-resolution AFM structure of DNA G-wires in aqueous solution. Nat. Commun. 9, 1959 (2018). https://doi.org/10.1038/s41467-018-04016-y
Note: The original article featured above was published as Open Access under a Creative Commons license. The data shown here are reused under fair use of the original article, which can be accessed through the article link above.
