AFM Systems
AFM Accessories
Learning
Contact Us
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
Researchers gained insight into the role of the enzyme PICH in DNA disentanglement through AFM images of molecular-level structure. They found PICH promoted positive DNA supercoiling that later helped in its disentanglement by another enzyme.
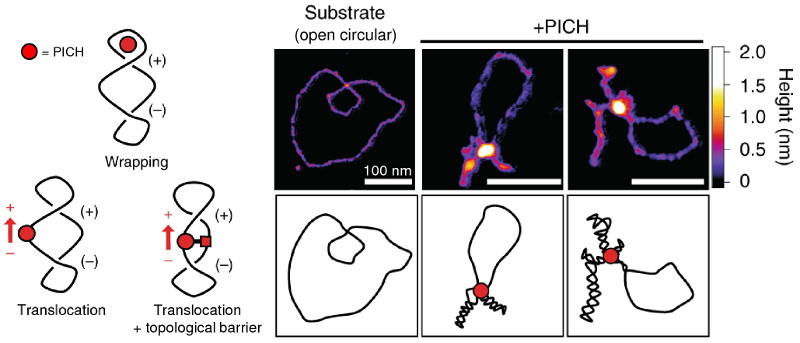
The long, helical nature of DNA molecules means that sister chromatids—a pair of soon-to-be individual chromosomes—are intricately entangled and intertwined. This intertwining can cause formation of thousands of ultrafine bridges, which must be resolved before cell division begins. Although the enzyme PICH (Plk1-interacting checkpoint helicase) is known to coat the ultrafine bridges, its role in disentanglement is not well understood.
While investigating this issue, a team of researchers at Danish and French universities identified an enzymatic reaction in which PICH induced positive DNA supercoiling, or overwinding. Moreover, the positive supercoiling actually assisted in subsequent DNA disentanglement by other enzymes (type II topoisomerases).
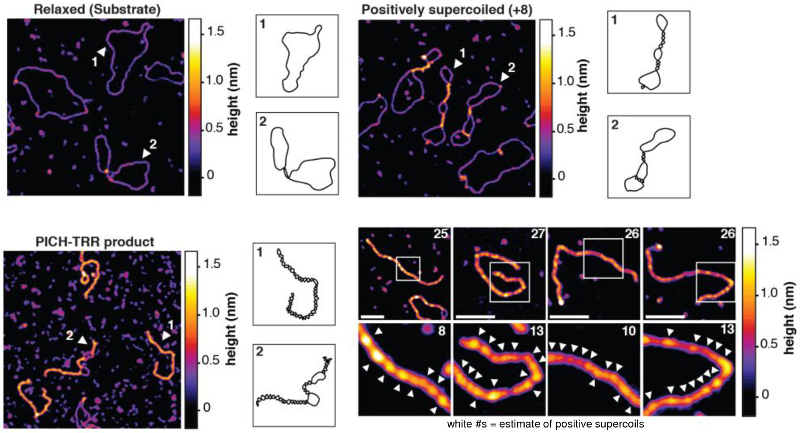
AFM images of molecular-level structure revealed that in the presence of the multi-subunit TRR complex containing type IA topoisomerase IIIα (Top3A), Rmi1, and Rmi24, PICH promoted extremely high levels of positive supercoiling. This activity had previously been considered unique to another class of enzymes called reverse gyrases. The team also proposed models to explain the supercoiling mechanism that were consistent with the observed DNA conformations.
The results advance our understanding of how DNA motor proteins make use of superhelicity to drive cellular processes. As such, they could provide insight into the molecular-level causes of tumor formation, pathological embryonic development, and other mechanisms.
Topography images of relaxed and supercoiled markers and post-reaction products were acquired in ambient conditions on a Cypher AFM with tapping mode. Images with 512x512 pixels were acquired at line scan rates up to 2 Hz, or less than 5 minutes per image. Combined with Cypher’s outstanding spatial resolution, these parameters meant images could be acquired relatively quickly yet still show molecular structure in detail.
Citation: A. Bizard, J.-F. Allemand, T. Hassenkam et al., PICH and TOP3A cooperate to induce positive DNA supercoiling. Nat. Struct. Mol. Biol. 26, 267 (2019). https://doi.org/10.1038/s41594-019-0201-6
Note: The data shown here are reused under fair use from the original article, which can be accessed through the article link above.
